

Research Article - International Research Journal of Biotechnology ( 2021) Volume 11, Issue 4
Received: 24-Nov-2020 Published: 30-Oct-2021
Climate change causes increasing the salinity crisis of arable land in the world. It is crucial to develop salt tolerant genotypes that can maintain optimum yield levels. Therefore, the aim of this study was to discover prospective genotypes tolerant to salinity by using Microsatellites/ Simple Sequence Repeats (SSR) primers. Selection for salinity tolerant genotypes of rice based on phenotypic performance alone is less reliable and will delay progress in breeding. Recent advents of molecular markers have been used in selecting salt tolerant rice genotypes. A total of 13 Simple Sequence Repeats (SSR) primers were used to identify salinity tolerance in 30 rice genotypes. A total number of 39 alleles were detected. The number of alleles per locus ranged from 3 to 6 with an average of 4.3. The Polymorphism Information Content (PIC) values ranged from 0.17 to 0.70 with an average of 0.51. The average gene diversity over all SSR loci for the 30 genotypes was 0.56, ranging from 0.18 to 0.74. The “Unweighted Pair Group Method of Arithmetic Means (UPGMA)” dendrogram constructed from Nei’s (1983) genetic distance produced three main distinct clusters among the 30 rice genotypes. Clusters are formed based on lower genetic distance between the genotypes. Cluster I consisted of only the salt tolerant check Pokkali, cluster II was made up of susceptible genotypes, while cluster III comprises of mostly moderately tolerant and tolerant genotypes. Molecular characterization and genetic diversity assessment is an important component in order to guide breeders in the choice of parents for a breeding programme.
Salinity tolerance, SSR primers, Rice (Oryza sativa), Polymorphism information content, Cluster analysis.
Rice (Oryza sativa) is one of the most important world food crops, serving as the staple food for over one-third of the world’s population (Nejad et al., 2010). Rice production is affected by many biotic and abiotic stresses throughout the world. Among these, abiotic stress alone contributes up to 50% of the total yield losses. The most fundamental of all abiotic stresses is salinity of soil/water which is considered prevalent limiting factor in rice production (Ren et al., 2010). Worldwide, 20% of all irrigated land is salt-affected (45 million out of 230 million ha), with soils in the arid and semiarid regions having excessive concentrations of soluble salts, which adversely affect plant growth (Aref and Rad, 2012). Salt stress is a major constraint to cereal production worldwide (Tuteja, 2012), it is therefore imperative to develop salt tolerant genotypes as a strategy to increase rice production in salinity-prone coastal areas. Rice yield in salt-affected land is significantly reduced with an estimation of 30–50% yield losses annually (Enyard et al., 2005). Due to natural and human interferences, the arable land is continuously transforming into saline, which will eventually lead to an overwhelming global effects and consequently will result in up to 50% land loss by 2050 (Saha et al., 2010, Hasanuzzaman et al., 2013). The conventional methods of plant selection for salt tolerance are not easy because of the large environmental effects and the low narrow sense heritability for this trait. Deoxyribonucleic acid (DNA) is the genetic material that contains the clue to identify the potentiality and diversity among different plant germplasms (Semagn et al. 2006). Molecular markers provide information that can help to define the distinctiveness of germplasms, rank them according to the number of close relative and assign their phylogenetic positions. Simple sequence repeats (SSR) or microsatellite markers have been identified to be ideal for creating genetic maps (Islam, 2004; Niones, 2004), assisting selection (Bhuiyan, 2005) and studying genetic diversity in germplasms. SSR markers are more popular in rice because they are highly informative, mostly monolocus, co-dominant, easy to analyze and cost effective (Chambers and Avoy, 2000). The present investigation was undertaken to genetically characterize genotypes using SSR primers that can serve as promising germplasm resources for breeders in selecting salt tolerant genotypes.
Plant materials
Thirty rice genotypes (modern released varieties, mutated lines and landraces) including salt tolerant (Pokkali) and salt susceptible (IR29) genotype, which were used as controls. Table 1 shows the selected genotypes for this study and were collected from the AfricaRice centre at the International Institute for Tropical Agriculture (IITA), Ibadan and Federal University of Agriculture Abeokuta (FUNAAB). The experiment was done at the Bioscience centre, IITA (Latitude 3° 54̍ N and longitude 7° 30̍ W).
| No. | Genotype | Pedigree | Country of origin | Specie | Response to stress |
|---|---|---|---|---|---|
| V1 | CK73 | - | Guinea | O. sativa | Salinity tolerant |
| V2 | FARO 44 (SPI 690233) |
SIPI 661044/SIPI 651020 | Nigeria | O. sativa | Blast resistant |
| V3 | FARO 52 (WITA 4) |
11975/IR 13146-45-2-3 | Senegal | Drought and iron toxicity tolerant | |
| V4 | FARO60 (NERICA – L19) | TOG5681/3*IR64 | Senegal | Interspecific | Blast, drought and iron toxicity resistance |
| V5 | FARO61 (NERICA L – 34) |
TOG5681/4*IR64 | Senegal | Interspecific | Drought and iron toxicity resistant |
| V6 | ITA212 (FARO 35) |
BG90-2*4/TE TEP | Nigeria | O. sativa | Salinity Tolerant |
| V7 | ITA 222 (FARO 36) |
MAHSURI/RPCB-2B-849 | Nigeria | O. sativa | Salinity tolerant |
| V8 | ITA306 | TOX 494-3-6-9-6/TOX 711//BG 6812 | Nigeria | O. sativa | Unknown |
| V9 | IR64 | IR 5657-33-2-1/IR 2061-465-1-5-5 | Philippines | O. sativa | Poor soil; Low input |
| V10 | IR72 | IR 19661-9-2-3-3/IR 15795-199-3-3/IR 9129-209-2-2-2-1 | Philippines | O. sativa | Unknown |
| V11 | NERICA L7 | TOG5681/3*IR64 | Senegal | Interspecific | Drought, cold and iron toxicity resistant |
| V12 | NERICA L8 | TOG5681/3*IR64 | Senegal | Interspecific | Drought, cold and iron toxicity resistant |
| V13 | NERICA L12- | TOG5681/3*IR64 | Senegal | Interspecific | Drought, cold and iron toxicity resistant |
| V114 | NERICA L53 | IR64/TOG5681//4*IR64 | Senegal | Interspecific | Drought cold and iron toxicity resistant |
| V15 | NERICA L42 | TOG5681/4*IR64 | Senegal | Interspecific | Drought, salt,cold and iron toxicity resistant |
| V16 | NERICA L45 | TOG5681/5*IR64 | Senegal | Interspecific | Drought, cold and iron toxicity resistant |
| V17 | NERICA L48 | IR64/TOG5681//4*IR64 | Senegal | Interspecific | Drought, cold and iron toxicity resistant |
| V18 V19 V20 |
NERICA L50 NERICA L54 OG0315 |
IR64/TOG5681//4*IR64 IR64/TOG5681//4*IR64 FUNAABOR-1-0M |
Senegal Senegal Nigeria |
Interspecific Interspecific O. Sativa |
Drought and iron toxicity resistant Drought, cold and iron toxicity resistant Drought resistant |
| V21 | OG250315 | FUNAABOR-1-25M | Nigeria | O. sativa | Drought resistant |
| V22 | OG300315 | FUNAABOR-1-30M | Nigeria | O. sativa | Drought resistant |
| V23 | OW0315 | FUNAABOR-2-0M | Nigeria | O. sativa | Unknown |
| V24 | OW100315 | FUNAABOR-2-10M | Nigeria | O. sativa | Unknown |
| V25 | OW250315 | FUNAABOR-2-25M | Nigeria | O. sativa | Unknown |
| V26 | ROK 5 | SR26/Pa WELLINGTON | Sierra Leone | Salinity tolerant | |
| V27 | ROK24 | SIAM 25/3*MALUNJA | Sierra Leone | Iron toxicity tolerant | |
| V28 | TOG 5681 | Landrace | Nigeria | O. glaberrima | |
| V29 | IR29 (Susceptible check) | IR833-6-2-1-1////IR1561-149-1//4*IR24/O. nivara | Philippines | Resistant to biotic stress; blast, tungro, gall midge. | |
| V30 | Pokkali (Tolerant check) |
IRIS 10-17834 9 (Land race) | India | O.glaberrima | Salinity resistant |
Genomic DNA isolation and SSR marker analysis
Total genomic DNA was extracted from young leaves using modified CTAB method (Nguyen et al., 2001). Thirteen SSR markers Covering 3 chromosomes of rice, were selected from the Genome Databases, Rice Genes Microsatellite Markers. These primer sequences were synthesized by MWG Biotech Inc Germany. Polymerase chain reactions (PCR) were carried out in PTC (A Programmable Thermal Cycler). The volume of the reaction mixture was 15 μL which consisted of 2.0 μL DNA sample and 13 μL Master mix (MWG Biotech Inc). The temperature cycles were programmed as 94°C for 5 min, followed by denaturation at 94°C for 1 min, annealing 55°C for 1 min, and polymerization at 72°C for 2 min for 35 cycles and additional temperature of 72°C for 7 min for final extension and 4°C for cooling until electrophoresis.
A 300 ml 0.5 X TBE (electrophoresis buffer) was taken in a flask, 4.5 g of agarose was added to it. The mixture was then cooked for 5 minutes in an oven to dissolve it. The gel was left at room temperature to cool down at tolerable level; the gel was then poured into gel mold carefully. Meanwhile two combs were placed on the gel, the gel polymerized within 30 minutes. The gel was submerged into 0.5 X TBE buffer in the gel tank. The combs were then removed from the gel tank. The PCR product was mixed with 3 μl of 2 X gel loading dye (MWG Biotech Inc). Eight microliter of the mixture was taken by the micro-pipette, loaded slowly per well on the gel. The molecular weight marker [100 bp (DNA ladder), 4 μl] was loaded at the first well on the gel. The tank was covered and all connections were checked. Electrophoresis machine was switched on at 120 volts for 1-1.30 hr. The separation process was monitored by the migration of the dyes in the loading buffer. When the bromophenol blue dye had reached about three-fourths of the gel length, the electrophoresis was stopped. Before electrophoresis, the gel mixture was stained with ethidium bromide solution.
The genetic diversity parameters such as major allele frequency, polymorphic information content (PIC) values which indicates the ability to distinguish between genotypes for each primer combination, heterozygosity and alleles per locus were computed using PowerMarker v. 3.25 (Liu and Muse, 2005). DARwin v. 6.0.13 (Perrier and Jacquemoud- Collet, 2006) was employed to generate genetic distance (GD) matrix followed by phylogeny reconstruction using unweighted neighbour joining (Nei et al., 1983).
Polymorphism information content was calculated using the formula:
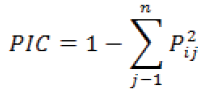
Where,
Pij = is the frequency of the jth allele for the ith marker, and summed over n alleles.
Microsatellites are among the most widely used DNA marker for many purposes such as diversity, genome mapping, varietal identification, etc. (Rashid et al, 2018). Genetic diversity assessment of modern varieties and traditional landraces are essential components in germplasms characterization and conservation to identify potential parents. A total of 39 alleles were detected at the loci of 9 microsatellite markers across 30 rice genotypes. The number of alleles per locus generated by each marker varied from 3.0 – 6.0 alleles, with an average of 4.3 alleles per locus. The number of alleles per locus (4.3) detected in this study was very much consistent with Pervaiz et al., 2010 and Rahman et al 2012 who found an average of 4.4 and 4.18 alleles per locus, respectively. Gholizadeh and Navabpour (2014) detected 3.53 alleles per SSR locus using 29 genotypes, which is lower than that reported in this study. The highest number of alleles was detected in the loci RM336, RM493 and RM3412 while the lowest number was detected in loci RM510, RM1287 and RM10745. In contrast, the number of alleles present in the current study was lower than the average number of alleles reported by Mizan et al., (2015) and Prathepha (2012) who reported an average of 8.44 and 11.85 alleles per locus. The variability in the number of alleles detected per locus might be due to the use of diverse genotypes and the selection of different SSR primers with scorable alleles. The average mean of allele frequency is 0.55, which suggests that on average 55% of the 30 genotypes shared a common major allele at any given locus. A moderate level of diversity exists among nine loci across 30 genotypes with an average of 56%. Figure 1 shows images of amplified fragments produced by primer RM336, RM341 and RM493. Primer RM336 on chromosome 7 detected 6 alleles and RM 3412 and RM493 both on chromosome 1 (saltol region of the rice genome) detected 5 and 6 alleles, respectively, suggesting that these markers could be used for molecular screening in selecting genotypes for salinity tolerance. The Polymorphic Information Content (PIC) values of a marker reflects marker allele diversity and frequency among genotypes. The PIC value in this study ranged from 0.17 (RM10745) to 0.70 (RM336) with an average of 0.51 per locus. Gholizadeh and Navabpour (2014) reported a PIC value ranging from 0.07 to 0.71 with an average of 0.45 using 30 SSR markers. RM336 had the highest PIC value followed by RM3412, RM493 and RM585. Therefore, SSR marker, RM336 was found to be superior for the analysis of genetic diversity in this study. Titov et al., (2009) used three primers in genotypic screening of rice for salt tolerance out of the three primers only RM336 distinguish nine out of 11 genotypes used and two as susceptible. Bhuiyan (2005) identified 158 tolerant individuals in the F2 and F3 population of BRRI Dhan 28 X PSBRc88 using the marker RM493. Niones (2004) reported five SSR markers (RM1287, RM8094, RM3412, RM493 and RM140) and two expressed sequence tag (EST) markers (CP3970 and CP6224) were linked to Saltol QTL on chromosome 1. It was also observed that Saltol locus is probably located within a region consisting of RM8094, RM3412, RM49 (Tables 2 and 3).
| Locus Name | Sequence | Motif | Chr | Ann. Temp (°C) |
|---|---|---|---|---|
| RM493 | F - TAGCTCCAACAGGATCGACC R – GTACGTAAACGCGGAAGGTG |
(CTT)9 | 1 | 56 |
| RM562 | F -CACAACCCACAAACAGCAAG R - CTTCCCCCAAAGTTTTAGCC |
(AAG)13 | 1 | 53 |
| RM510 | F - AACCGGATTAGTTTCTCGCC R - TGAGGACGACGAGCAGATTC |
(GA)15 | 6 | 55 |
| RM585 | F - CAGTCTTGCTCCGTTTGTTG R - CTGTGACTGACTTGGTCATAGG |
(TC)45 | 6 | 55 |
| RM140 | F - TGCCTCTTCCCTGGCTCCCCTG R - GGCATGCCGAATGAAATGCATG |
(CT)12 | 1 | 58 |
| RM336 | F - CTTAACAGAGAAACGGCATCG R - GCTGGTTTGTTTCAGGTTCG |
(CTT)18 | 7 | 55 |
| RM8115 | F - TATATAGTAAATTTGTTTGGTGTAGG R - ACAGATGGATATTATAAGAAGTAACA |
(TA)18 | 1 | 50 |
| RM8094 | F - AAGTTTGTACACATCGTATACA R - CGCGACCAGTACTACTACTA |
(AT)31 | 1 | 55 |
| RM10772 | F - GCACACCATGCAAATCAATGC R - CAGAAACCTCATCTCCACCTTCC |
(CTT)16 | 1 | 55 |
| RM10745 | F - TGACGAATTGACACACCGAGTACG R - ACTTCACCGTCGGCAACATGG |
(TATG)9 | 1 | 55 |
| RM1287 | F - GTGAAGAAAGCATGGTAAATG R - CTCAGCTTGCTTGTGGTTAG |
(AG)17 | 1 | 55 |
| RM3412 | F - AAAGCAGGTTTTCCTCCTCC R - CCCATGTGCAATGTGTCTTC |
(CT)17 | 1 | 55 |
| RM10764 | F - AGATGTCGCCTGATCTTGCATCG R - GATCGACCAGGTTGCATTAACAGC |
(AT)28 | 1 | 55 |
| Markers | Major allele frequency | Allele number | Gene diversity | PIC |
|---|---|---|---|---|
| RM336 | 0.33 | 6.00 | 0.74 | 0.70 |
| RM3412 | 0.36 | 5.00 | 0.72 | 0.67 |
| RM493 | 0.43 | 6.00 | 0.71 | 0.66 |
| RM510 | 0.53 | 3.00 | 0.55 | 0.45 |
| RM562 | 0.50 | 4.00 | 0.63 | 0.57 |
| RM585 | 0.53 | 5.00 | 0.64 | 0.60 |
| RM1287 | 0.86 | 3.00 | 0.23 | 0.22 |
| RM10772 | 0.50 | 4.00 | 0.62 | 0.55 |
| RM10745 | 0.90 | 3.00 | 0.18 | 0.17 |
| Mean | 0.55 | 4.33 | 0.56 | 0.51 |
The genetic similarity were ted from the data of coefficients (Table 4). The similarity matrix was used to determine the level of relatedness among the studied genotypes. The highest genetic dissimilarity was between IR29 and ITA222 (86%). The lowest genetic dissimilarity among the genotypes was between NERICA L7 and NERICA L12 (0%), followed by ITA306 and FARO 52 (7%), OG300315 and OG250315 (7%), OG300315 and OG0315 (7%), OW250315 and OG250315 (7%). The clustering in Figure 2 presents an overview about genetic diversity and it represents the genetic similarity matrix among the studied genotypes derived from the microsatellite analysis shown in Table 4. Assessment of genetic diversity is very important in plant breeding as it provides the basis for selection. The SSR markers played an important role in determining germplasm diversity in rice (Yu et al., 2005). Genetic diversity is commonly measured by genetic distance or genetic similarity, both of which imply that there are either differences or similarities at the genetic level (Weir, 1990). The results of this study indicated that SSR analysis could be a better method to study the genetic diversity in rice. Thirty rice genotypes were divided into 3 distinct clusters. Cluster I contains the salt tolerant check this might be because none of the other 29 genotypes produced similar alleles as Pokkali (Figure 2). Cluster II was divided into two sub-groups where the first group was made up of the highly susceptible (the salt sensitive check IR29 and Rok5) and susceptible genotypes. Cluster III had the moderately tolerant, tolerant and highly tolerant geneotypes. The tolerant (ITA222, FARO 44, ITA212, FARO 52 and ITA306) and highly tolerant (NERICA L8, NERICA L54, NAERICA L50, NERICA L48, FARO 61 and IR64) genotypes were included in sub-groups II and III, respectively, based on the genetic distance indices. The clustering revealed an interesting fact that genotypes with derivatives of genetically similar type cluster together. Based on the above result genotypes were grouped in the same cluster due to lower genetic distance of the gene types and higher similarity.
| OTU | V1 | V2 | V3 | V4 | V5 | V29 | V9 | V10 | V6 | V7 | V8 | V13 | V15 | V16 | V17 | V18 | V14 | V19 | V11 | V12 | V22 | V21 | V20 | V23 | V24 | V25 | V30 | V26 | V27 | V28 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1 | 0.00 | |||||||||||||||||||||||||||||
| V2 | 0.64 | 0.00 | ||||||||||||||||||||||||||||
| V3 | 0.57 | 0.29 | 0.00 | |||||||||||||||||||||||||||
| V4 | 0.57 | 0.57 | 0.43 | 0.00 | ||||||||||||||||||||||||||
| V5 | 0.50 | 0.29 | 0.14 | 0.36 | 0.00 | |||||||||||||||||||||||||
| V29 | 0.64 | 0.64 | 0.71 | 0.79 | 0.79 | 0.00 | ||||||||||||||||||||||||
| V9 | 0.57 | 0.29 | 0.21 | 0.36 | 0.07 | 0.79 | 0.00 | |||||||||||||||||||||||
| V10 | 0.42 | 0.71 | 0.64 | 0.64 | 0.57 | 0.50 | 0.64 | 0.00 | ||||||||||||||||||||||
| V6 | 0.50 | 0.43 | 0.14 | 0.36 | 0.29 | 0.64 | 0.36 | 0.57 | 0.00 | |||||||||||||||||||||
| V7 | 0.57 | 0.57 | 0.29 | 0.36 | 0.29 | 0.86 | 0.36 | 0.64 | 0.36 | 0.00 | ||||||||||||||||||||
| V8 | 0.57 | 0.36 | 0.07 | 0.43 | 0.14 | 0.79 | 0.21 | 0.64 | 0.21 | 0.29 | 0.00 | |||||||||||||||||||
| V13 | 0.29 | 0.50 | 0.36 | 0.50 | 0.43 | 0.57 | 0.50 | 0.57 | 0.29 | 0.57 | 0.43 | 0.00 | ||||||||||||||||||
| V15 | 0.57 | 0.57 | 0.29 | 0.50 | 0.43 | 0.50 | 0.50 | 0.43 | 0.21 | 0.36 | 0.36 | 0.43 | 0.00 | |||||||||||||||||
| V16 | 0.43 | 0.43 | 0.50 | 0.29 | 0.36 | 0.79 | 0.43 | 0.57 | 0.43 | 0.43 | 0.50 | 0.50 | 0.57 | 0.00 | ||||||||||||||||
| V17 | 0.57 | 0.29 | 0.29 | 0.43 | 0.14 | 0.71 | 0.07 | 0.64 | 0.43 | 0.43 | 0.29 | 0.43 | 0.57 | 0.43 | 0.00 | |||||||||||||||
| V18 | 0.64 | 0.21 | 0.14 | 0.43 | 0.14 | 0.71 | 0.14 | 0.71 | 0.29 | 0.43 | 0.21 | 0.43 | 0.43 | 0.50 | 0.21 | 0.00 | ||||||||||||||
| V14 | 0.43 | 0.50 | 0.50 | 0.29 | 0.43 | 0.79 | 0.50 | 0.57 | 0.43 | 0.43 | 0.50 | 0.50 | 0.57 | 0.07 | 0.50 | 0.57 | 0.00 | |||||||||||||
| V19 | 0.57 | 0.43 | 0.36 | 0.50 | 0.29 | 0.71 | 0.21 | 0.64 | 0.50 | 0.36 | 0.36 | 0.50 | 0.50 | 0.57 | 0.14 | 0.36 | 0.50 | 0.00 | ||||||||||||
| V11 | 0.29 | 0.50 | 0.36 | 0.50 | 0.43 | 0.57 | 0.50 | 0.57 | 0.29 | 0.57 | 0.43 | 0.00 | 0.43 | 0.50 | 0.43 | 0.42 | 0.50 | 0.50 | 0.00 | |||||||||||
| V12 | 0.57 | 0.29 | 0.21 | 0.57 | 0.21 | 0.71 | 0.29 | 0.57 | 0.36 | 0.36 | 0.29 | 0.43 | 0.36 | 0.50 | 0.29 | 0.21 | 0.57 | 0.29 | 0.43 | 0.00 | ||||||||||
| V22 | 0.50 | 0.64 | 0.50 | 0.50 | 0.50 | 0.57 | 0.43 | 0.36 | 0.43 | 0.43 | 0.50 | 0.64 | 0.21 | 0.57 | 0.50 | 0.57 | 0.57 | 0.43 | 0.64 | 0.57 | 0.00 | |||||||||
| V21 | 0.50 | 0.64 | 0.57 | 0.57 | 0.57 | 0.50 | 0.50 | 0.36 | 0.50 | 0.50 | 0.57 | 0.57 | 0.29 | 0.57 | 0.43 | 0.64 | 0.57 | 0.36 | 0.57 | 0.57 | 0.07 | 0.00 | ||||||||
| V20 | 0.50 | 0.64 | 0.50 | 0.50 | 0.50 | 0.57 | 0.50 | 0.36 | 0.43 | 0.43 | 0.50 | 0.64 | 0.21 | 0.57 | 0.57 | 0.50 | 0.57 | 0.50 | 0.64 | 0.57 | 0.07 | 0.14 | 0.00 | |||||||
| V23 | 0.43 | 0.71 | 0.64 | 0.64 | 0.64 | 0.50 | 0.64 | 0.43 | 0.57 | 0.64 | 0.64 | 0.71 | 0.43 | 0.57 | 0.64 | 0.64 | 0.57 | 0.64 | 0.71 | 0.71 | 0.29 | 0.29 | 0.21 | 0.00 | ||||||
| V24 | 0.57 | 0.64 | 0.57 | 0.64 | 0.64 | 0.50 | 0.64 | 0.43 | 0.50 | 0.57 | 0.50 | 0.57 | 0.29 | 0.64 | 0.57 | 0.57 | 0.64 | 0.50 | 0.57 | 0.57 | 0.21 | 0.14 | 0.14 | 0.29 | 0.00 | |||||
| V25 | 0.50 | 0.64 | 0.57 | 0.57 | 0.57 | 0.50 | 0.57 | 0.36 | 0.50 | 0.50 | 0.57 | 0.57 | 0.29 | 0.57 | 0.50 | 0.57 | 0.57 | 0.43 | 0.57 | 0.57 | 0.14 | 0.07 | 0.07 | 0.21 | 0.07 | 0.00 | ||||
| V30 | 0.50 | 0.50 | 0.50 | 0.43 | 0.43 | 0.43 | 0.50 | 0.57 | 0.64 | 0.57 | 0.50 | 0.64 | 0.71 | 0.64 | 0.50 | 0.57 | 0.64 | 0.50 | 0.64 | 0.50 | 0.71 | 0.71 | 0.71 | 0.64 | 0.79 | 0.71 | 0.00 | |||
| V26 | 0.64 | 0.43 | 0.50 | 0.64 | 0.64 | 0.36 | 0.64 | 0.50 | 0.43 | 0.71 | 0.57 | 0.57 | 0.36 | 0.50 | 0.64 | 0.57 | 0.50 | 0.71 | 0.57 | 0.64 | 0.43 | 0.43 | 0.42 | 0.42 | 0.43 | 0.43 | 0.71 | 0.00 | ||
| V27 | 0.50 | 0.50 | 0.36 | 0.43 | 0.43 | 0.71 | 0.50 | 0.64 | 0.29 | 0.43 | 0.43 | 0.36 | 0.43 | 0.21 | 0.50 | 0.43 | 0.21 | 0.57 | 0.36 | 0.43 | 0.64 | 0.63 | 0.64 | 0.64 | 0.64 | 0.64 | 0.71 | 0.50 | 0.00 | |
| V28 | 0.43 | 0.50 | 0.36 | 0.50 | 0.50 | 0.57 | 0.57 | 0.50 | 0.21 | 0.43 | 0.43 | 0.36 | 0.29 | 0.50 | 0.50 | 0.50 | 0.50 | 0.43 | 0.36 | 0.36 | 0.36 | 0.29 | 0.35 | 0.50 | 0.29 | 0.29 | 0.64 | 0.50 | 0.43 | 0.00 |
In summary, the result from this study outlined an implication for engineering salt tolerance genotypes based on microsatellite clusters; the SSR markers used in this study could not classify the selected genotypes into different salttolerant categories. However, we observed an association between a representative set of studied genotypes with known salt tolerant SSR markers (RM336, RM3412 and RM493). These markers may be useful in screening for salt tolerance in rice genotypes. Futhermore, the association of these markers with salt tolerant genes/quantitative trait loci needs to be confirmed. The SSR marker analysis in this study is capable of detecting major gene locus for plant breeders to develop new salt tolerant genotypes. The identified salt tolerant rice genotypes of the present study could be further tested in saline areas to observe yield potential for developing high yielding genotypes suitable for saline areas.